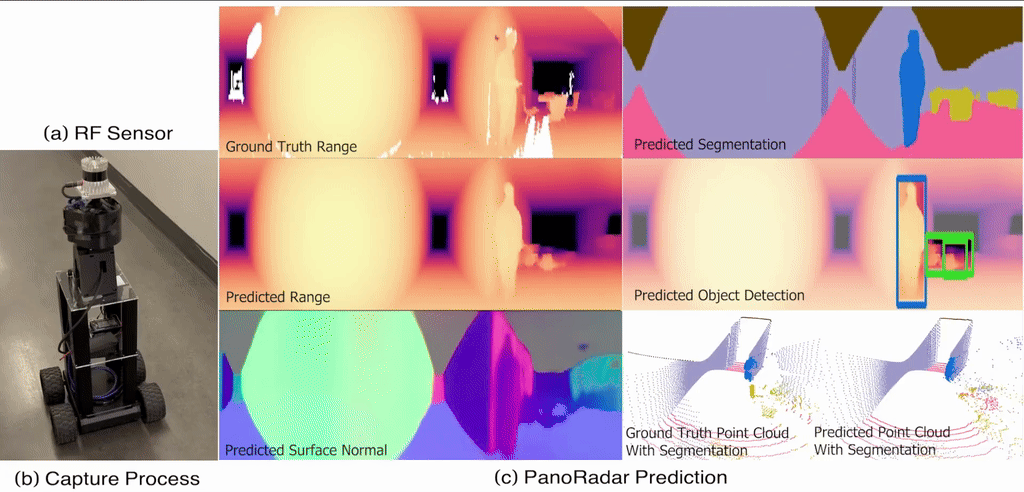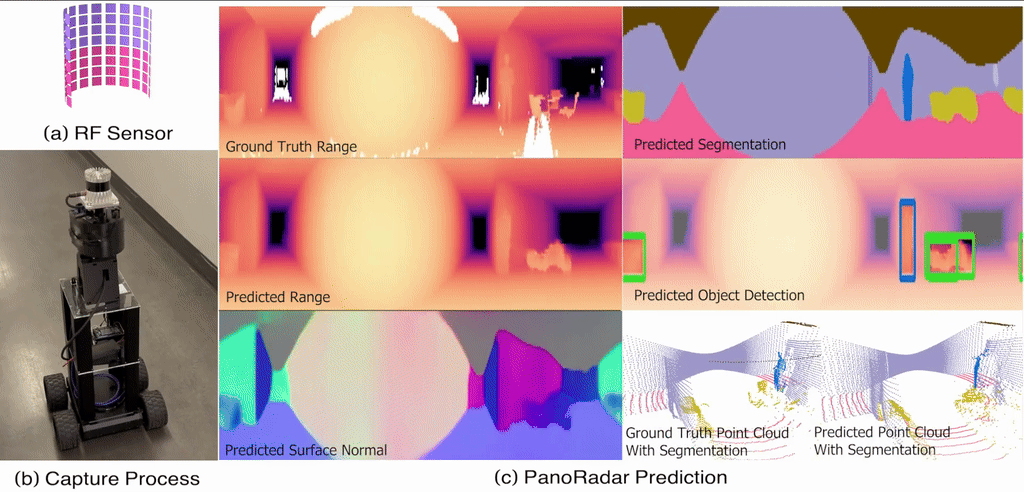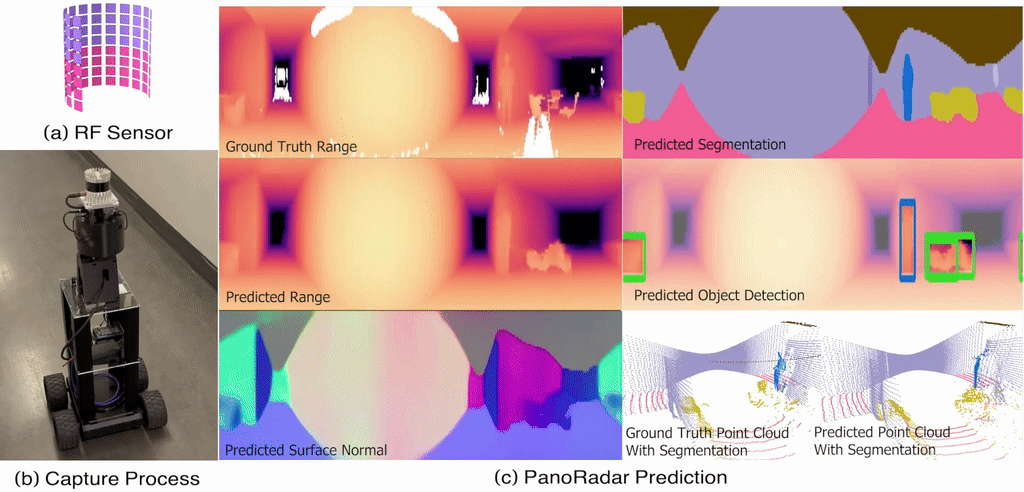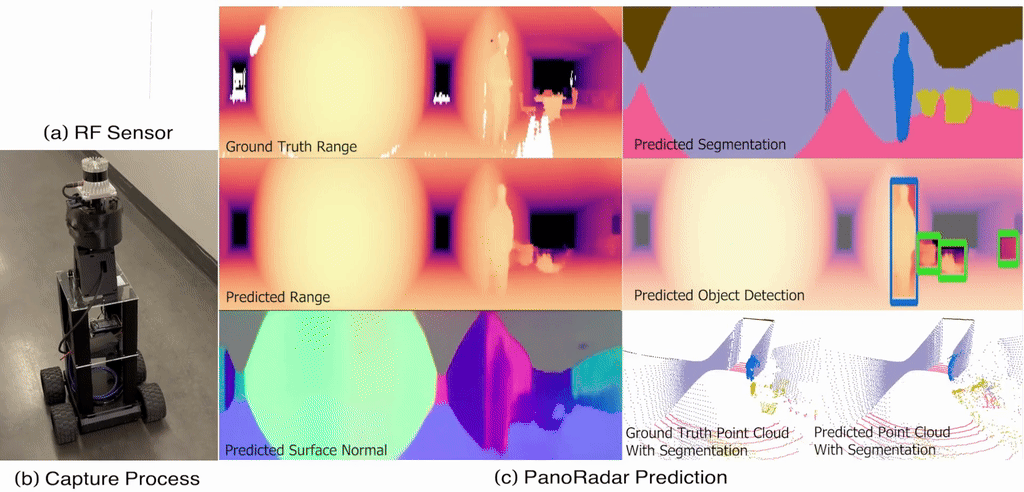
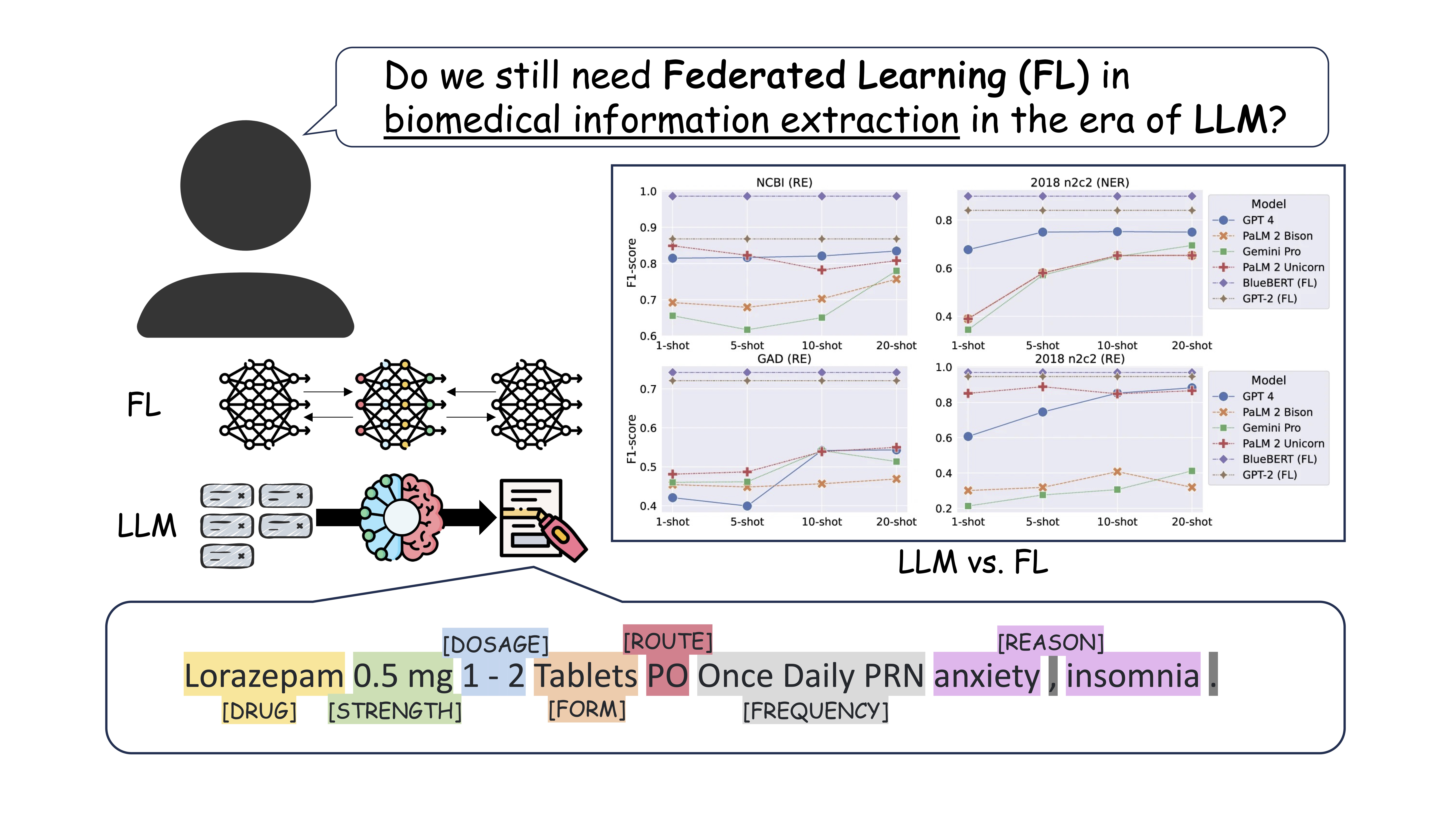
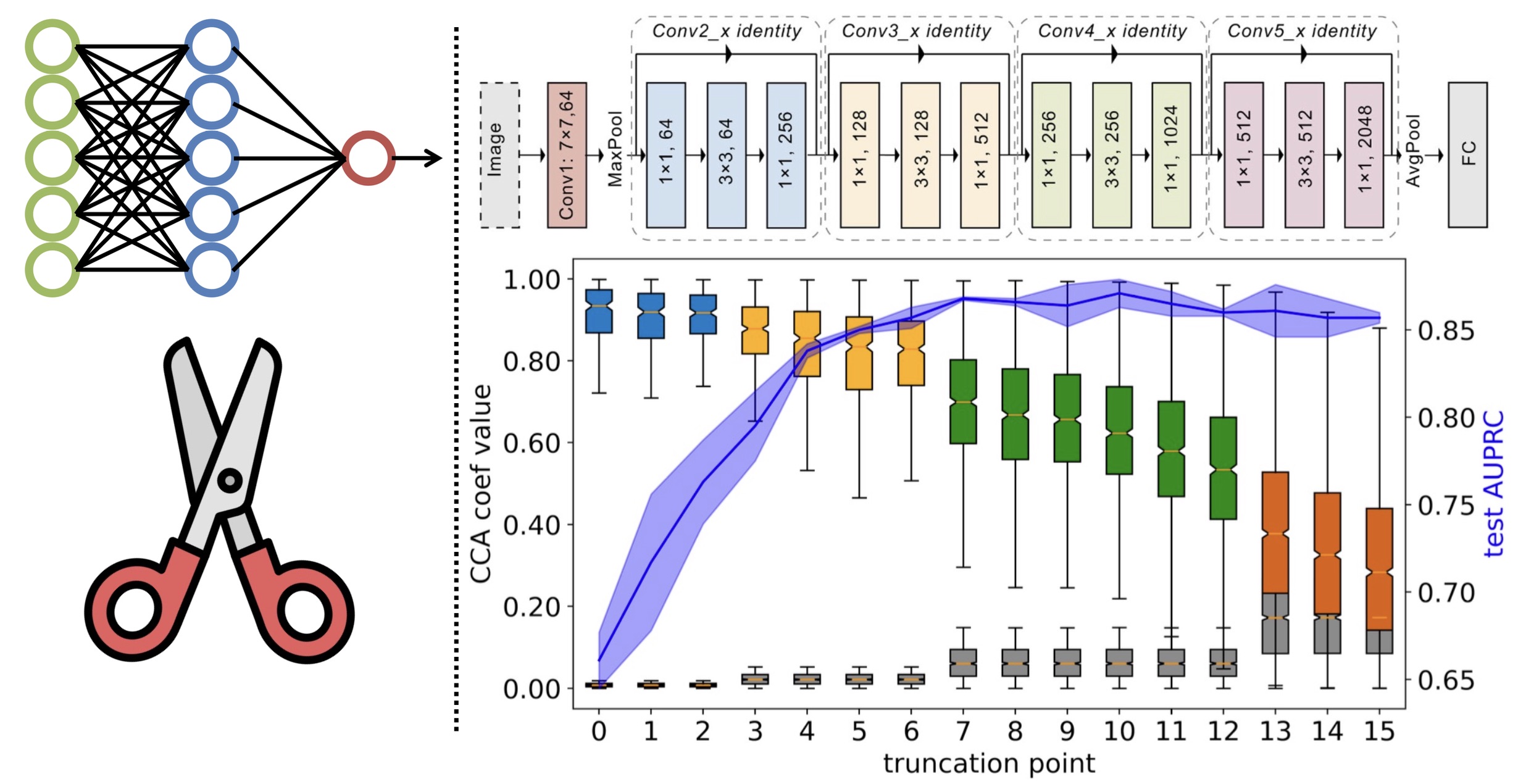
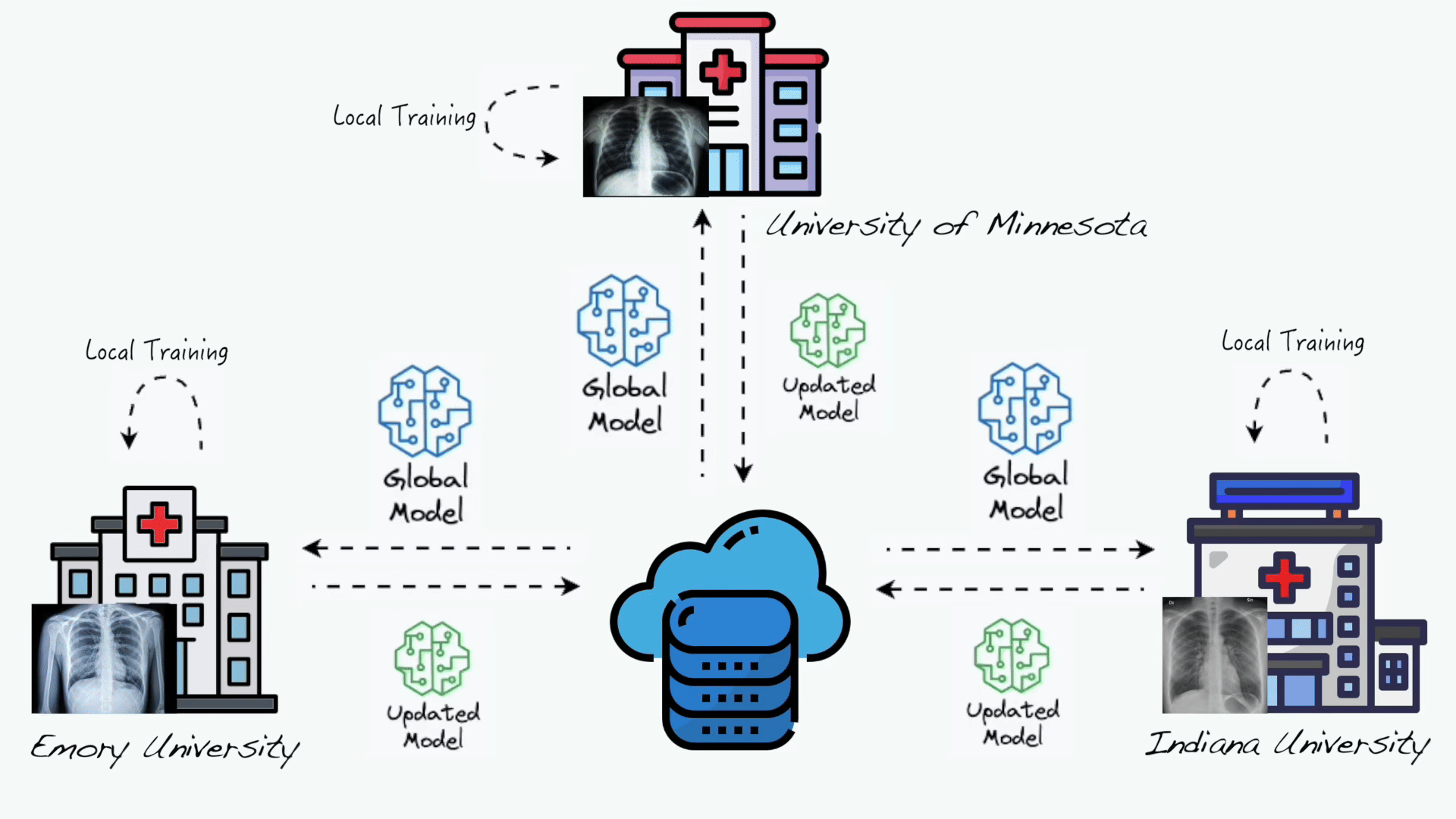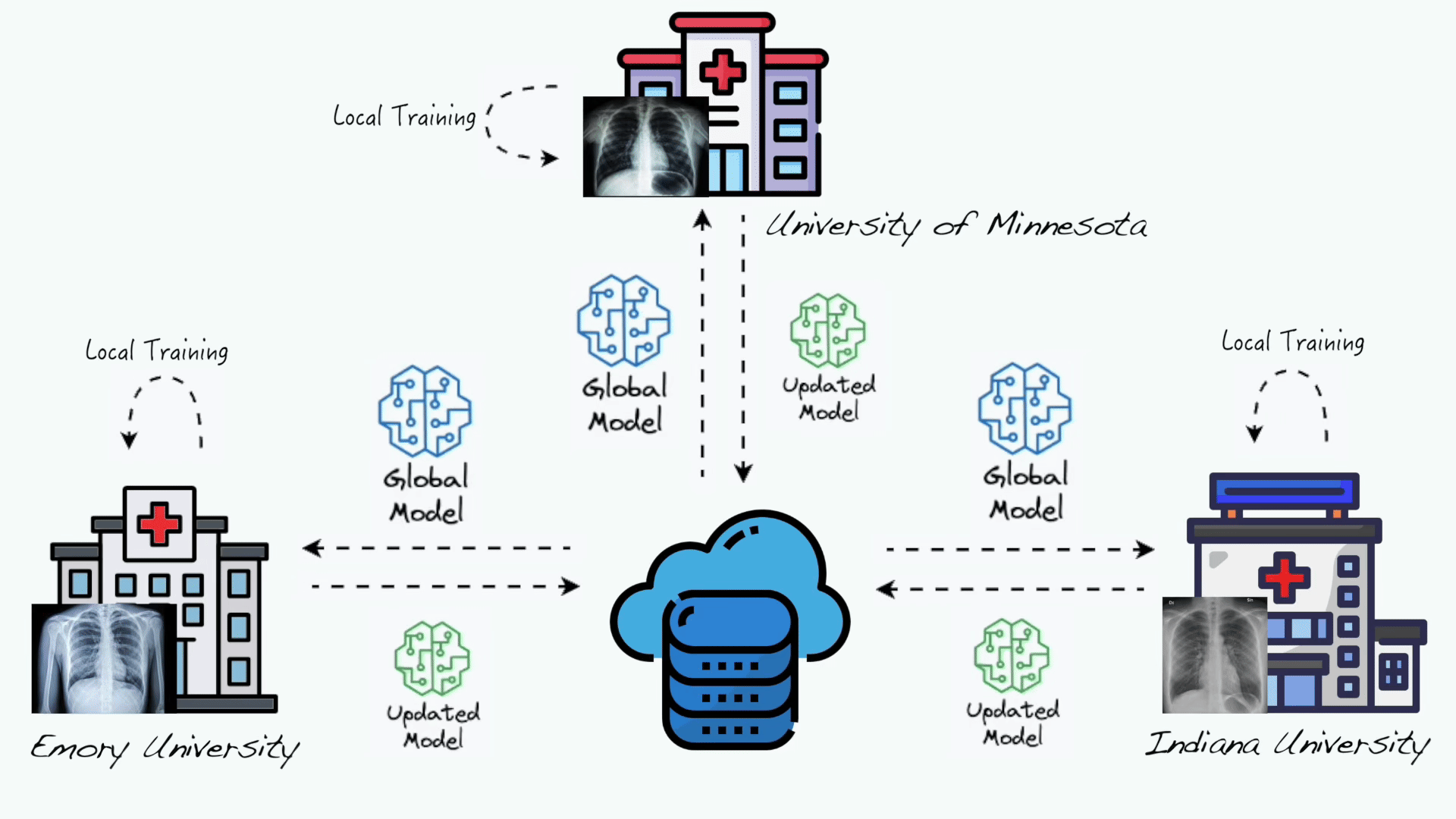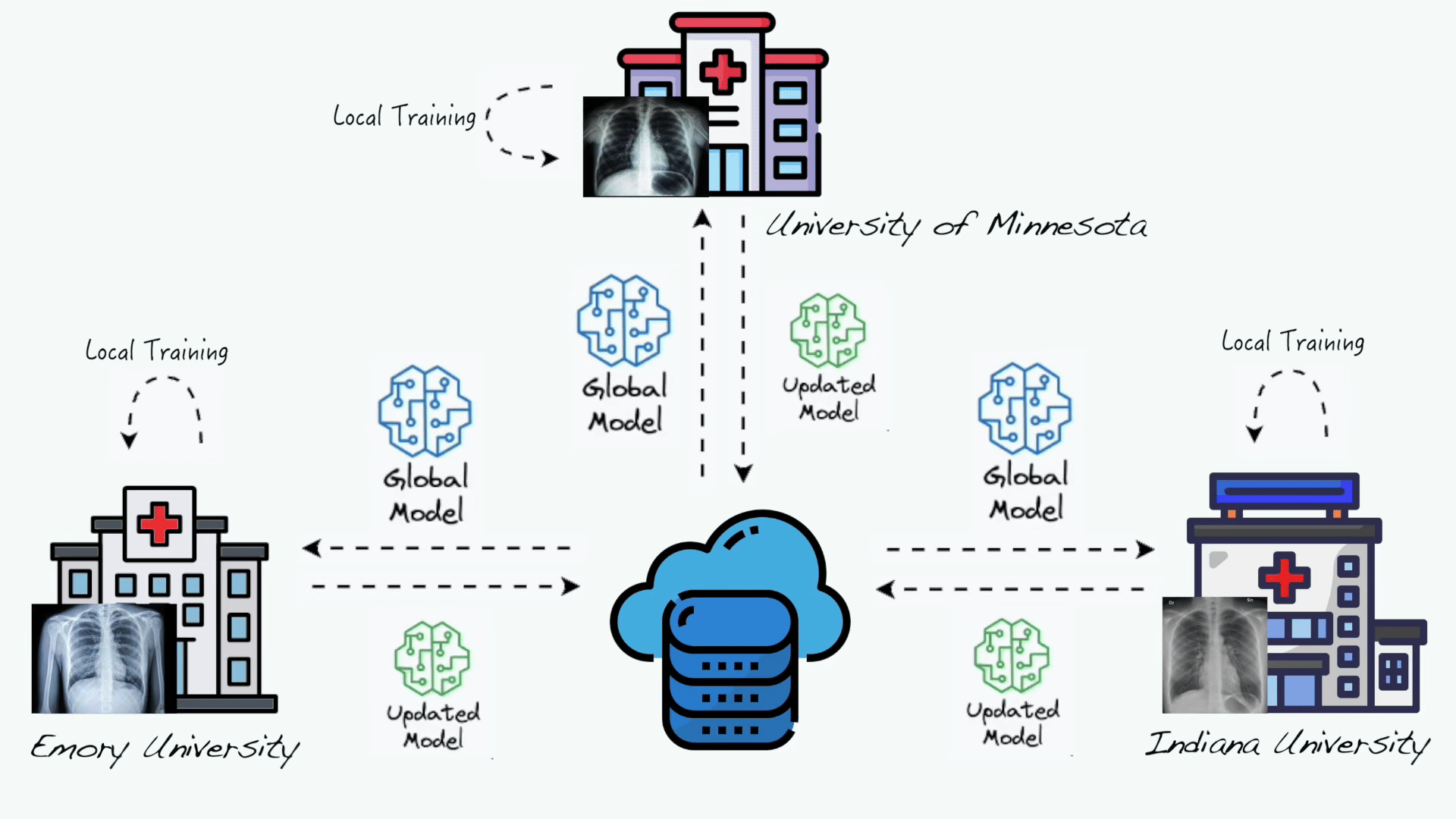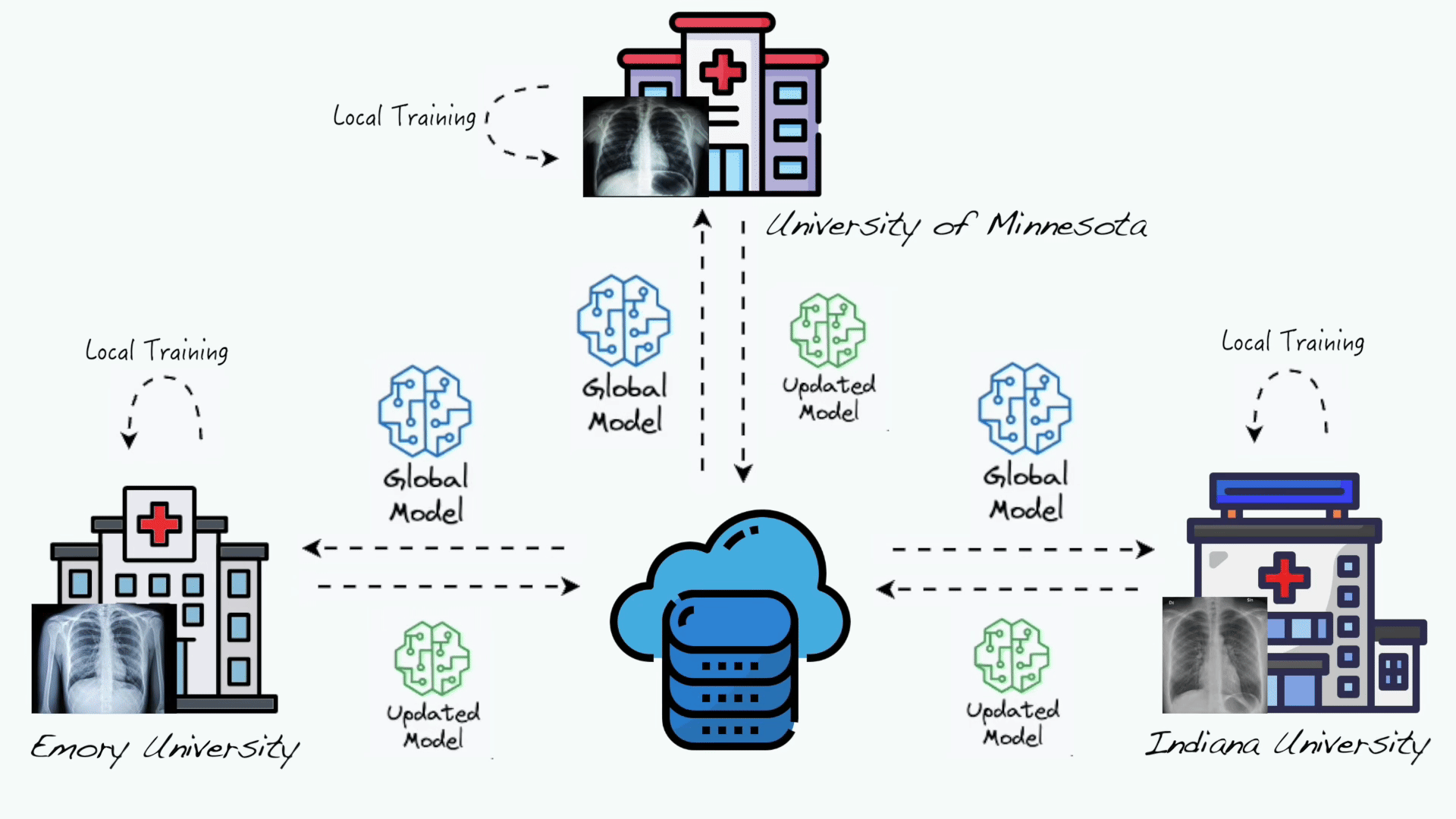
Gaoxiang Luo
(I go by Leon)
Ph.D. CandidateGroup of Learning, Optimization, Vision, Healthcare, and X (GLOVEX)
Department of Computer Science and Engineering
University of Minnesota, Twin Cities
luo00042{at}umn[dot]edu
Keller Hall 6-210Hey there! I’m Gaoxiang Luo and I’m a 2nd-year Ph.D. candidate at the University of Minnesota, Twin Cities, advised by Prof. Ju Sun and Prof. Aryan Deshwal. I’m broadly interested in generative models. My pursuit of a doctorate is funded by Zscaler and DSI-MnDRIVE award.
Previously: I received my M.S. in Computer and Information Science from University of Pennsylvania. I was a research intern at Endeavor AI, Zscaler, and Cisco Research and a recipient of Google CSRMP.
News
| Feb, 2026 | Our paper Flow Matching for Multimodal Distributions has been accepted to CVPR 2026! |
|---|---|
| Feb, 2026 | The qualitative study of our open-access tool MetaMate has been accepted to CHI 2026 (Poster)! |
| Dec, 2025 | I led a team of 4 and won the AgentDS Hackathon overall 1st place 🥇! |
| Sep, 2025 | Thanks to Synthesis Company (YC S24) for supporting server hosting for MetaMate! |
| Aug, 2025 | A paper accepted to the EMNLP 2025 main conference! |
Latest Posts
| Jun 20, 2024 | Import or Upgrade Third Party Python Packages in Snowflake Streamlit and Notebook without UDFs |
|---|---|
| May 20, 2024 | VC Methods |
| Aug 11, 2023 | Outshift | Interning with Cisco Research |